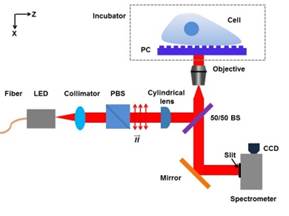
 Photonic Crystal-Enhanced Microscopy (PCEM) is a powerful multimode imaging approach that combines label-free (LF) and enhanced-fluorescence (EF) imaging modalities for quantification and visualization of the interactions between cells and their substrate. The PCEM is designed to utilize the properties of photonic crystal (PC) biosensor surface that is designed to produce bandwidth optical resonance for two distinct purposes. First, the resonant coupling condition of the PC is modified by cell attachment, and the PCEM is capable of producing images of spatial distribution of cell attachment with sufficient resolution to quantify variation in the strength of attachment within individual cells. Second, the PC surface enables efficient coupling to laser illumination, resulting in the ability to enhance the output of fluorescent molecules in close proximity to PC through a technique called “PC-Enhanced Fluorescence.” The PCEM will be an enabling tool for a broad range of life science research and pharmaceutical screening applications that require characterization of the interaction between extracellular matrix materials and cell membrane-expressed proteins in response to cell growth, pro-apoptotic stimuli, forced differentiation protocols, chemotaxis, and proliferation. PCEM offers the ability to perform assays with a limited number of available cells for applications involving primary cells or stem cells.
Photonic Crystal-Enhanced Microscopy (PCEM) is a powerful multimode imaging approach that combines label-free (LF) and enhanced-fluorescence (EF) imaging modalities for quantification and visualization of the interactions between cells and their substrate. The PCEM is designed to utilize the properties of photonic crystal (PC) biosensor surface that is designed to produce bandwidth optical resonance for two distinct purposes. First, the resonant coupling condition of the PC is modified by cell attachment, and the PCEM is capable of producing images of spatial distribution of cell attachment with sufficient resolution to quantify variation in the strength of attachment within individual cells. Second, the PC surface enables efficient coupling to laser illumination, resulting in the ability to enhance the output of fluorescent molecules in close proximity to PC through a technique called “PC-Enhanced Fluorescence.” The PCEM will be an enabling tool for a broad range of life science research and pharmaceutical screening applications that require characterization of the interaction between extracellular matrix materials and cell membrane-expressed proteins in response to cell growth, pro-apoptotic stimuli, forced differentiation protocols, chemotaxis, and proliferation. PCEM offers the ability to perform assays with a limited number of available cells for applications involving primary cells or stem cells.
Label-free imaging
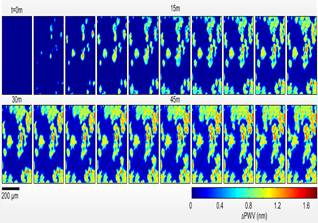
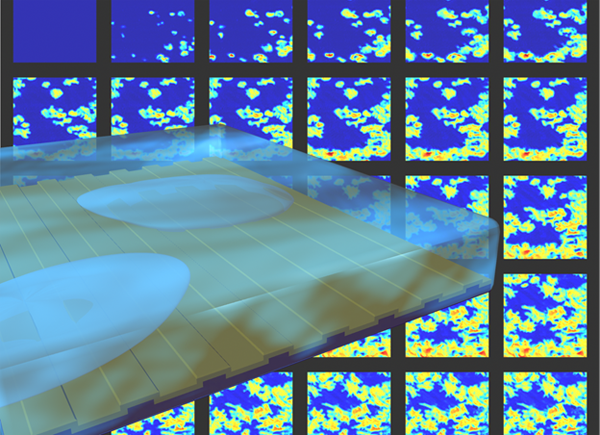
Photonics Crystal-Enhanced Microscopy (PCEM) is capable of quantifying cell-surface interactions with spatial resolution sufficient for monitoring intra-cell attachment distribution and temporal resolution sufficient for generating time-lapse “movies” of cell-surface interactions during various processes. Long-term monitoring of cell attachment over substantial time scales (hours to days) is allowed without interruption of the culture environment. Attachment and chemotaxis processes of dental stem cells are clearly observed. Single-cell movement and filopodial extension were easily identifiable, yielding significant potential for the future investigation of numerous cellular processes including tumor cell metastasis and stem cell differentiation.
The innovation of PCEM is based on the resonance properties of the PC biosensor. When the PC is illuminated with a broadband light source, the resonant wavelength is modulated by the addition of biomaterial upon the PC surface, resulting in a shift to a higher wavelength. The electromagnetic standing wave that is generated at the PC surface during resonant light coupling inhibits lateral propagation, thus enabling neighboring regions on the PC surface to display a distinct resonant wavelength that is determined only by the density of biomaterial attached at that precise location. By measuring the resonant peak wavelength value (PWV) on a pixel-by-pixel basis over a PC surface, an image of cell attachment density may be recorded.
The attachment of murcine dental stem cells to a PC surface was studied over a period of two hours. (Shown in Fig. 2.) Cells are observed attaching to the treated surface, with initial attachment characterized by small, round PWV shifts, consistent with spherical cells exiting suspension. As time progresses, average cell diameter increases, and membrane boundaries become more irregular as cellular processes begin to extend from cell bodies.
Enhanced-fluorescence imaging
While label-free cell attachment imaging provides important information about cell behavior and has many benefits, it can be augmented substantially through combination with fluorescence imaging. The ability to gather both types of images simultaneously would allow study of the effects of adding a stimulus (such as a drug) to a population of biosensor-attached cells and to dynamically measure surface attachment and the distribution of fluorophore-tagged structures inside and outside the cell. Currently no cell analysis tool exists that is capable of simultaneously performing label-free imaging and enhanced fluorescence imaging.
The most important aspect of fluorescence enhancement in PCEM is that the enhancement effect is restricted to a small evanescent field region above the PC surface. The PC can be either illuminated at an “on-resonance” coupling angle to produce the enhancement effect or in an “off-resonance” manner that results in no surface-enhanced electric field. By comparing an “on-resonance” PCEF image to an “off-resonance” PCEF image and comparing the intensities of the two images, it is possible to determine which fluorescent emitters are located within the evanescent field region and which are outside the evanescent field. Because the evanescent field strength decreases exponentially as one moves away from the PC surface, the on/off resonance ratio can be used to accurately determine the distance between a fluorescent source. This method can be used as an additional tool for studying the interaction of cell membranes with surfaces.
Label-free PCEM for Nanoparticles
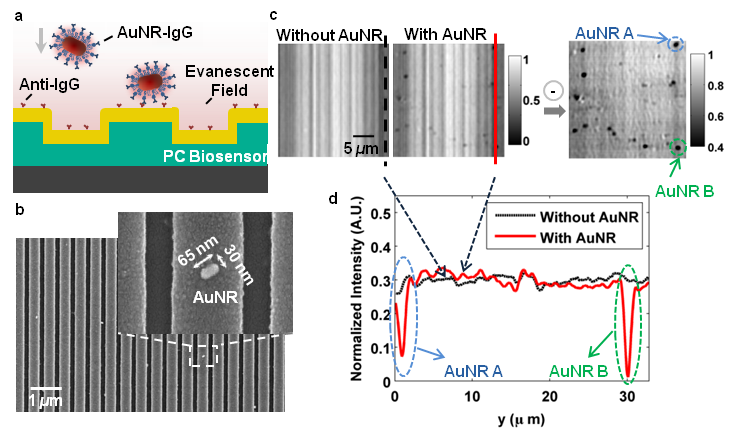
The Nanosensors Group has demonstrated a label-free biosensor imaging approach that utilizes a photonic crystal surface to detect surface attachment of individual dielectric and metal nanoparticles through measurement of localized shifts in the resonant wavelength and resonant reflection magnitude from the photonic crystal.
Using a microscopy-based approach to scan the photonic crystal resonant reflection properties with 0.6 µm spatial resolution, we show that metal nanoparticles attached to the biosensor surface with strong absorption at the resonant wavelength induce a highly localized reduction in reflection efficiency and are able to be detected by modulation of the resonant wavelength. Experimental demonstrations of single-nanoparticle imaging are supported by Finite Difference Time Domain (FDTD) computer simulations. By matching the metal nanoparticle plasmon resonant frequency to the PC resonance, we substantially increase the sensitivity of the imaging approach to the detection of individual surface adsorbed nanoparticles (size of 65×30×30 nm^3).
The ability to image surface adsorption of individual nanoparticles offers a route to single molecule biosensing in which the particles can be functionalized with specific recognition molecules and utilized as tags. Therefore, our work lays the foundation for the use of nanoparticles as tags for biomolecular assays performed upon a PC surface and measured by a PCEM instrument. We envision the development of the sandwich-style assays in which the PC surface is prepared with capture molecules for low-concentration analytes (such as cancer biomarkers), where nanoparticles tags prepared with secondary antibodies can decorate the captured analyte. Such an approach may enable multiplexed detection of analytes with single molecule resolution in the future.

Related Papers
- “Photonic crystal enhanced microscopy for imaging of live cell adhesion,” W. Chen, K.D. Long, M. Lu, V. Chaudhery, H. Yu, J.S. Choi, J. Polans, Y. Zhuo, B.A. Harley, and B.T. Cunningham, Analyst, Vol. 138, No. 20, p. 5886-5894, 2013.
- “Single nano-particle detection using photonics crystal-enhanced microscopy,” Y. Zhuo, H. Hu, W. Chen, M. Lu, L. Tian, H. Yu, K.D. Long, E. Chow, W.P. King, S. Singamaneni, and B.T. Cunningham, Analyst, Vol. 139, No. 5, p. 1007-1015, 2014.
- “Enhanced live-cell imaging via photonics crystal-enhanced fluorescence microscopy,” W. Chen, K.D. Long, H. Yu, Y. Tan, J.S. Choi, B.A. Harley, and B.T. Cunningham, Analyst, Vol. 139, No. 22, p. 5955-5964, 2014.