CHAMPAIGN, Ill. — A fast, inexpensive yet sensitive technique to detect cancer markers is bringing researchers closer to a “liquid biopsy – a test using a small sample of blood or serum to detect cancer, rather than the invasive tissue sampling routinely used for diagnosis.
Researchers at the University of Illinois developed a method to capture and count cancer-associated microRNAs, or tiny bits of messenger molecules that are exuded from cells and can be detected in blood or serum, with single-molecule resolution. The team published its results in the Proceedings of the National Academy of Science.
“Cancer cells contain gene mutations that enable them to proliferate out of control and to evade the immune system, and some of those mutations turn up in microRNAs,” said study leader Brian Cunningham, an Illinois professor of electrical and computer engineering. Cunningham also directs the Holonyak Micro and Nanotechnology Lab at Illinois.
“There are specific microRNA molecules whose presence and concentration is known to be related to the presence and aggressiveness of specific types of cancer, so they are known as biomarkers that can be the target molecule for a diagnostic test,” he said.
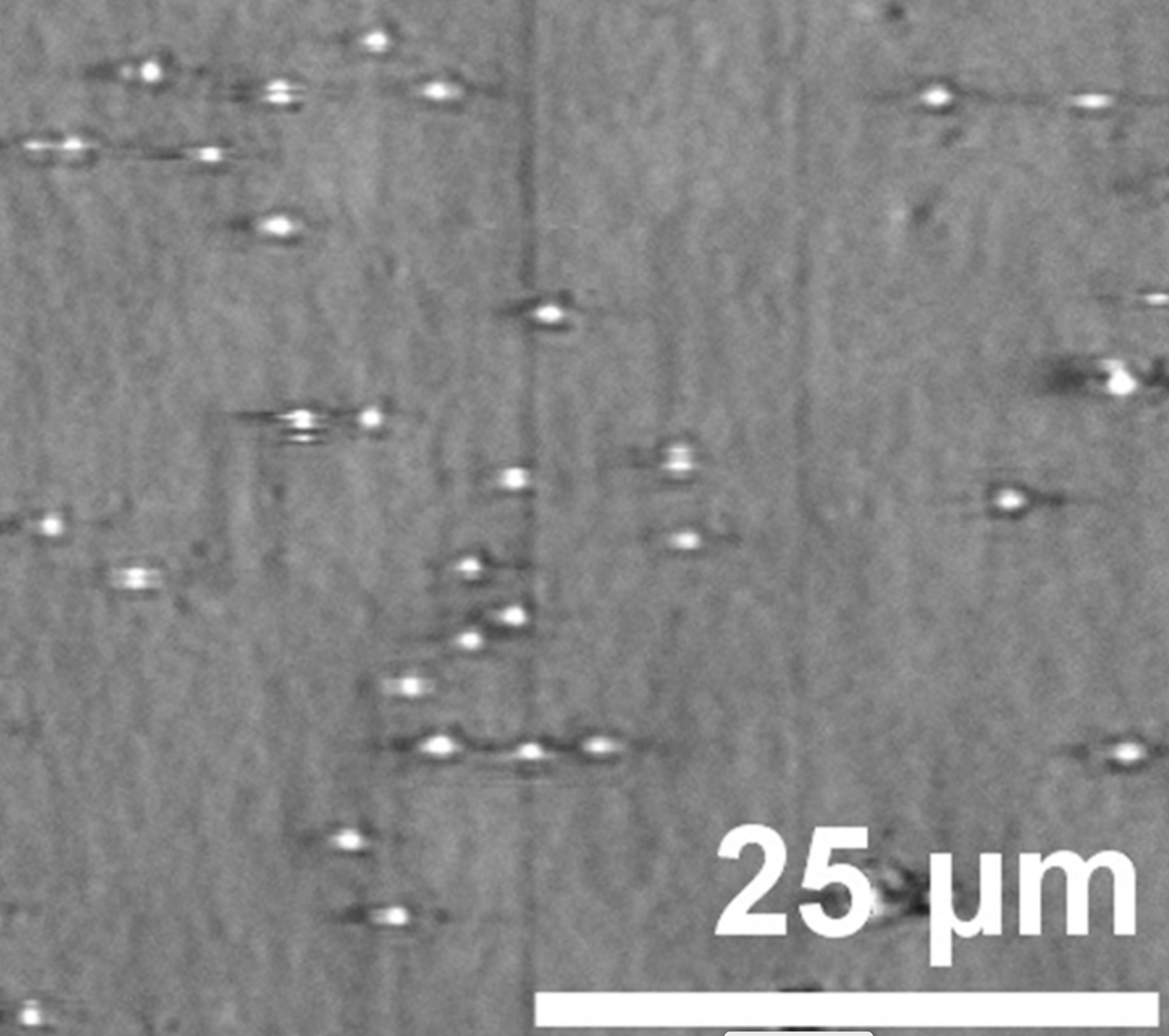
Cunningham’s group developed a technique named Photonic Resonator Absorption Microscopy to capture and count microRNA biomarkers. In collaboration with professor Manish Kohli at the Moffitt Cancer Center in Florida, they tested PRAM on two microRNAs that are known markers for prostate cancer.
They found it was sensitive enough to detect small amounts that would be present in a patient’s serum, yet also selective enough to detect the marker among a cocktail of molecules that also would be present in serum.
“One of the main challenges of biosensing is to maintain sensitivity and selectivity at the same time,” said Nantao Li, a graduate student and co-first author. “You want it to be sensitive enough to detect very small amounts, but you don’t want it to pick up every RNA in the blood. You want this specific sequence to be your target.”
PRAM achieves both qualities by combining a molecular probe and a photonic crystal sensor. The probe very specifically pairs to a designated microRNA and has a protective cap that comes off when it finds and binds to the target biomarker. The exposed end of the probe can then bind to the sensor, producing a signal visible through a microscope.
Each individual probe that binds sends a separate signal that the researchers can count. This means researchers are able to detect much smaller amounts than traditional methods like fluorescence, which need to exceed a certain threshold to emit a measurable signal. Being able to count each biomarker also carries the added benefit of allowing researchers to monitor changes in the concentration of the biomarker over time.
“With PRAM, we squirt a sample into a solution and get a readout within two hours,” said postdoctoral researcher Taylor Canady, a co-first author of the study. “Other technologies that produce single-molecule readouts require extra processing and additional steps, and they require a day or more of waiting. PRAM seems like something that could be much more feasible clinically. In addition, by using an optical signal instead of fluorescence, we could one day build a miniaturized device that doesn’t need a trained laboratory technician.”
The PRAM approach could be adapted to different microRNAs or other biomarkers, the researchers say, and is compatible with existing microscope platforms.
“This approach makes the idea of performing a ‘liquid biopsy’ for low-concentration cancer-related molecules a step closer to reality,” Cunningham said. “This advance demonstrates that it is possible to have an inexpensive and routine method that is sensitive enough to require only a droplet of blood. The results of the test might tell a physician whether a regimen of chemotherapy is working, whether a person’s cancer is developing a new mutation that would make it resistant to a drug, or whether a person who had been previously treated for cancer might be having a remission.”
The Carl R. Woese Institute for Genomic Biology at the U. of I. and the National Institutes of Health supported this work. Illinois chemistry professor Yi Lu and bioengineering professor Andrew Smith were coauthors of the work.
Editor’s note: To reach Brian Cunningham, call 217-265-6291; email bcunning@illinois.edu. The paper “Digital-resolution detection of microRNA with single-base selectivity by photonic resonator absorption microscopy” is available online. DOI: 10.1073/pnas.1904770116